A novel tetrameric PilZ domain structure from xanthomonads
Li, T.-N., Chin, K.-H., Fung, K.-M., Yang, M.-T., Wang, A.H.-J., Chou, S.-H.(2011) PLoS One 6: e22036-e22036
- PubMed: 21760949
- DOI: https://doi.org/10.1371/journal.pone.0022036
- Primary Citation of Related Structures:
3RQA - PubMed Abstract:
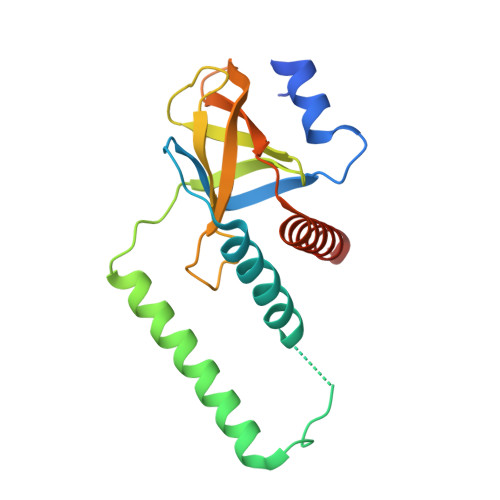
PilZ domain is one of the key receptors for the newly discovered secondary messenger molecule cyclic di-GMP (c-di-GMP). To date, several monomeric PilZ domain proteins have been identified. Some exhibit strong c-di-GMP binding activity, while others have barely detectable c-di-GMP binding activity and require an accessory protein such as FimX to indirectly respond to the c-di-GMP signal. We now report a novel tetrameric PilZ domain structure of XCC6012 from the plant pathogen Xanthomonas campestris pv. campestris (Xcc). It is one of the four PilZ domain proteins essential for Xcc pathogenicity. Although the monomer adopts a structure similar to those of the PilZ domains with very weak c-di-GMP binding activity, it is nevertheless interrupted in the middle by two extra long helices. Four XCC6012 proteins are thus self-assembled into a tetramer via the extra heptad repeat α3 helices to form a parallel four-stranded coiled-coil, which is further enclosed by two sets of inclined α2 and α4 helices. We further generated a series of XCC6012 variants and measured the unfolding temperatures and oligomeric states in order to investigate the nature of this novel tetramer. Discovery of this new PilZ domain architecture increases the complexity of c-di-GMP-mediated regulation.
Organizational Affiliation:
Institute of Biochemistry, National Chung-Hsing University, Taichung, Taiwan, Republic of China.